Samples, DNA extraction, and sequencing
Permission to analyze the Japanese wolf DNA was obtained for Jentink b (Leiden b): RMNH.MAM.39183, and Jentink c (Leiden c): RMNH. MAM.39181(52994) in Naturalis Biodiversity Center, Leiden, the Netherlands, and Jw284 (ZMB48817): ZMB_Mam_48817 in Museum für Naturkunde, Berlin, Germany. Six Japanese wolf samples from personal collections (Jw229, Jw255, Jw258, Jw269, Jw271, and Jw275) were analyzed with the permission of the owners. Blood or saliva samples for Japanese dogs were analyzed with the permission of the owners. Japanese Wolf DNAs were extracted and used in our previous mitochondrial DNA studies: Ishiguro et al. 2009 (Jw229, Jw255, and Jw258)22, Ishiguro et al. 2016 (Jw269 and Jw271)37, and Matsumura et al. 2021 (Jw275, Jw284, Leiden b, and Leiden c)25. The sample locations are listed in Supplementary Data 1. Bone powder (0.1 to 0.3 g) was obtained from the mandible (Jw229, Jw255, Jw258, Jw269) and Cranium (Jw271 and Jw275) specimens by using an electric drill after removal of the outer layers of bone by scraping with a sterile razor blade. Powders were also obtained from ventral nasal concha specimens (Jw284, Leiden b, and Leiden c) with a Multi-beads Shocker. According to methods described by Okumura et al. (1999)38, all bone powders were suspended in 10 ml of 0.5 M ethylenediamine tetraacetic acid (EDTA) at pH 7.0, and rotated for 24 hours for decalcification. This procedure was repeated several times until the supernatant became clear. The pellets of bone powder were collected by centrifugation, and the decalcification was repeated by washing with 10 ml of 0.5 M EDTA at pH 7.0 until the supernatant was clear. The samples were then treated with 5 ml of 0.5 M EDTA with proteinase K (300 μg/ml) and N-lauryl sarcosine (0.5%) for 24 hours. After centrifugation, the supernatant containing DNA was extracted twice with phenol, once with chloroform/phenol (1:1), and once with chloroform to remove protein. The supernatant was concentrated by using a Centricon 30 spin column (Amicon, Beverly, MA).
In recent years, Japanese dog breeding has become popular in Japan, and wide varieties of Shiba have been seen, including miniature Shiba and white or black Shiba (Shiba was initially light brown). However, what breeds were crossbred with Japanese dogs was unknown. Therefore, DNA extracted 26 years ago39 was used to avoid the effects of recent crossbreeding between Japanese dogs and other dog breeds for the eight modern dogs (Akita26, Akita27, Akita3, Kishu23, Kishu24, Shiba17, Shiba21, and Shiba60). Blood samples for these individuals were provided by the veterinary clinics with the permission of the owners. Peripheral blood leukocytes were suspended in a lysis buffer containing proteinase K (1 mg/ml). Genomic DNA was extracted twice with phenol, once with chloroform/isoamyl alcohol (24:1), then precipitated by ethanol.
Blood samples for two individuals of Shiba (Shiba_shiro and Shiba_kuro) were provided by the veterinary clinics with the permission of the owners. The saliva of an individuals of Shiba (Jm) was scrubbed by the owner with cotton swabs. DNAs of these three individuals of Shiba were extracted using a DNeasy Blood & Tissue Kit (Qiagen) following the manufacturer’s instructions. Paired-end (2 × 150 bp) sequencing was performed on the Illumina HiSeq X or NovaSeq 6000 platforms. The Institutional Animal Care and Use Committee of Anicom Specialty Medical Institute approved the animal protocols and procedures (No. 2020-02).
For Leiden b and Leiden c, the genome capture was performed using the SeqCap EZ Hybridization and Wash Kit (Roche, Basel, Switzerland), SeqCap EZ Accessory Kit v2 (Roche), SeqCap HE-Oligo Kit (Roche), and SeqCap EZ Pure Capture Bead Kit (Roche) following the manufacturer’s instructions for SeqCap EZ Library SR (Roche), with minor modifications. Briefly, Biotin-labeled genomic DNA fragments from Shiba were used as hybridization probes, instead of the SeqCap EZ library (Roche). Leiden b and Leiden c libraries were mixed with 135 ng of Biotin-labeled genomic DNA fragments and were hybridized at 47 °C for 72 h. Other procedures were performed in accordance with the manufacturer’s instructions. Total mapped reads are listed in Supplementary Data 1.
Extraction of SNPs and vcf file preparation
We downloaded sequencing data of 56 modern dogs, 23 modern gray wolves, four ancient dogs, five ancient canids, and six outgroup species from the database (Supplementary Data 2). Sequence reads from the genomic DNA libraries of nine Japanese wolves, eleven Japanese dogs (Supplementary Data 1) as well as 94 samples from the database (Supplementary Data 2) were trimmed to remove nucleotides with base qualities lower than 35 on average of a 150 bp read (sum of the base-calling error probability <0.05 in a 150 bp read) and adaptor sequences using CLC Genomics Workbench (https://www.qiagenbioinformatics.com/). The trimmed reads were mapped to the dog reference genome (CanFam3.1) using CLC Genomics Workbench. Reads showing high similarity ( > 90% in > 90% of read length) were mapped to the reference genome sequences to avoid mapping the low similarity reads. Reads mapped to more than one position were removed (“ignore” option for reads mapped to multiple positions) to prevent mapping to non-unique regions. The mapping data was exported in bam file format and sorted and indexed using samtools40. The duplicated reads in bam files were marked by the MarkDuplicates algorithm implemented in GATK v4.2 (https://gatk.broadinstitute.org/hc/en-us). We performed genotype calling on all individuals analyzed in this study using the HaplotypeCaller algorithm in GATK v4.2. Genotypes of all individuals were output as gvcf format (-ERC GVCF option). All gvcf files were combined into a single gvcf format file by the CombineGVCFs algorithm in GATK v4.2. The combined file was genotyped by the GenotypeGVCFs algorithm and filtered by Filtervcf in GATK v4.2 with parameters; –filter-expression “QD<2.0” –filter-name “QD2” –filter-expression “QUAL<30.0” –filter-name “QUAL30” –filter-expression “FS>200.0” –filter-name “FS200” –filter-expression “SOR>10.0” -filter-name “SOR10” –filter-expression “ReadPosRankSum<−20.0” –filter-name “ReadPosRankSum-20”.
Following the method of vonHoldt et al. (2010)10, we excluded the sample pairs that show IBS > 0.8 within groups (the same dog breeds or wolves from the same location). IBS values were obtained by the “—distance ibs” function of plink41. The IBS of two wolf pairs, Wolf_Chinese1-Wolf_Chinese2 and Wolf_Iberia1-Wolf_Iberia2, show higher IBS than the threshold (0.8). Therefore, we excluded Wolf_Chinese2 and Wolf_Iberia2 from all of our analyses.
To maximize the number of SNPs for analyses, we prepared datasets from the genotyped vcf file for each analysis by following filtering using vcftools42.
Dataset 1: PCA and ADMIXTURE using the Japanese wolf (excluding Leiden b, Leiden c, and a Honshu wolf) and modern samples (Fig. 1, Supplementary Figs. 4 and 5)
We removed four ancient dogs, five ancient canids, three Japanese wolves (Leiden b, Leiden c, and a Honshu wolf), two modern wolves (a Chinese wolf and an Iberian wolf), and three outgroup species (Andean fox, Dhole, and Ethiopian wolf) from the genotyped vcf file (Supplementary Data 2). We removed all sites with missing data. Then, we removed all indels, singleton, and doubleton sites to eliminate PCR and sequencing errors that may have occurred in one individual by minor allele frequency (MAF) filtering (MAF < 0.015). We extracted bi-allelic sites with coverage equal to or more than three in all individuals and with GQ values equal to or more than 20 in all individuals. Mutations due to DNA damage at both ends of fragments were less than 1% in Japanese wolves (Supplementary Fig. 1), therefore we can infer that mutations by DNA damage in the sequences of Japanese wolves were removed by this filtration. The final dataset consisted of 1,696,115 sites (97 individuals).
Dataset 2: PCA and ADMIXTURE using the Japanese wolf (including Leiden b, Leiden c, and a Honshu wolf) and modern samples (Supplementary Figs. 6 and 7)
We removed four ancient dogs, five ancient canids, two modern wolves (a Chinese wolf and an Iberian wolf), and three outgroup species (Andean fox, Dhole, and Ethiopian wolf) from the genotyped vcf file (Supplementary Data 2). We removed sites with missingness higher than 3% and minor allele frequency (MAF) < 0.04. We extracted bi-allelic sites with coverage equal to or more than three in all individuals and with GQ values equal to or more than eight in all individuals. The final dataset consisted of 342,931 sites (100 individuals).
Dataset 3: PCA using the Japanese wolf (excluding Leiden b, Leiden c, and a Honshu wolf), ancient dogs, ancient canids, and modern samples (Supplementary Fig. 9)
We removed two modern wolves (a Chinese wolf and an Iberian wolf), three Japanese wolves (Leiden b, Leiden c, and a Honshu wolf), and three outgroup species (Andean fox, Dhole, and Ethiopian wolf) from the genotyped vcf file (Supplementary Data 2). We removed all sites with missing data. Then, we removed all indels, singleton, and doubleton sites to eliminate PCR and sequencing errors that may have occurred in one individual by minor allele frequency filtering (MAF < 0.01). We extracted bi-allelic sites with coverage equal to or more than three in all individuals and with GQ values equal to or more than eight in all individuals. We extracted transversion sites to eliminate the effect of DNA damage in the ancient samples. The final dataset consisted of 100,588 sites (106 individuals).
Dataset 4: PCA and ADMIXTURE using the Japanese wolf (excluding Leiden b, and Leiden c), ancient canids, and modern samples (Supplementary Figs. 10–12)
We removed two modern wolves (a Chinese wolf and an Iberian wolf), and three outgroup species (Andean fox, Dhole, and Ethiopian wolf) from the genotyped vcf file (Supplementary Data 2). We modified this dataset to create a total of eight datasets, seven with the number of Japanese wolves ranging from 1 to 7 and one with only one Honshu wolf instead of Japanese wolves. We removed all sites with missing data. Then, we removed all indels and filtered by minor allele frequency filtering (MAF < 0.05). We extracted bi-allelic sites with coverage equal to or more than three in all individuals and with GQ values equal to or more than eight in all individuals. We extracted transversion sites to eliminate the effect of DNA damage in the ancient samples. The numbers of sites in the eight datasets are listed in Supplementary Fig. 12.
Dataset 5: f3, f4 statistics, and f4-ratio using the Japanese wolf (excluding Leiden b, Leiden c, and a Honshu wolf) and modern samples (Figs. 2B–D, 3A, C, Supplementary Figs. 25–29, 32−38)
We removed four ancient dogs, five ancient canids, three Japanese wolves (Leiden b, Leiden c, and a Honshu wolf), two modern wolves (a Chinese wolf and an Iberian wolf), and five outgroup species (Andean fox, Dhole, Ethiopian wolf, Golden Jackal, and African Golden Wolf) from the genotyped vcf file (Supplementary Data 2). We removed all sites with missing data. Then, we removed all indels, singleton, and doubleton sites to eliminate PCR and sequencing errors that may have occurred in one individual by minor allele frequency filtering (MAF < 0.01). We extracted bi-allelic sites with coverage equal to or more than three in all individuals and with GQ values equal to or more than 20 in all individuals. The final dataset consisted of 489,529 sites (95 individuals).
Dataset 6: f4 statistics and f4-ratio using the Japanese wolf (including Leiden b or Leiden c) and modern samples (Supplementary Fig. 8)
We removed four ancient dogs, five ancient canids, a Japanese wolf (a Honshu wolf), two modern wolves (a Chinese wolf and an Iberian wolf), and five outgroup species (Andean fox, Dhole, Ethiopian wolf, Golden Jackal, and African Golden Wolf) from the genotyped vcf file (Supplementary Data 2). We modified this dataset to create two datasets, one with Leiden b and the other with Leiden c. We removed all sites with missing data. Then, we removed all indels, singleton, and doubleton sites to eliminate PCR and sequencing errors that may have occurred in one individual by minor allele frequency filtering (MAF < 0.015). We extracted bi-allelic sites with coverage equal to or more than three in all individuals and with GQ values equal to or more than eight in all individuals. The final datasets with Leiden c and Leiden b consisted of 38,254 and 83,259 sites, respectively (70 individuals).
Dataset 7: Phylogenetic analyses using the Japanese wolf (excluding Leiden b, Leiden c, and a Honshu wolf) and modern samples (Supplementary Figs. 19–21, 31)
We removed four ancient dogs, five ancient canids, three Japanese wolves (Leiden b, Leiden c, and a Honshu wolf), and two modern wolves (a Chinese wolf and an Iberian wolf) from the genotyped vcf file (Supplementary Data 2). We removed all sites with missing data. Then, we removed all indels, singleton, and doubleton sites to eliminate PCR and sequencing errors that may have occurred in one individual by minor allele frequency filtering (MAF < 0.01). We extracted bi-allelic sites with coverage equal to or more than three in all individuals and with GQ values equal to or more than 20 in all individuals. The final dataset consisted of 2,065,002 sites (99 individuals).
Dataset 8: Phylogenetic analyses using the Japanese wolf (excluding Leiden b, Leiden c, and a Honshu wolf), African dogs, modern wolves, and outgroup samples (Supplementary Figs. 30, 31)
We removed four ancient dogs, five ancient canids, three Japanese wolves (Leiden b, Leiden c, and a Honshu wolf), five modern wolves (a Chinese wolf, an Iberian wolf, and three Middle East wolves), and dogs except for African dogs from the genotyped vcf file (Supplementary Data 2). We removed all sites with missing data. Then, we removed all indels, singleton, and doubleton sites to eliminate PCR and sequencing errors that may have occurred in one individual by minor allele frequency filtering (MAF < 0.03). We extracted bi-allelic sites with coverage equal to or more than three in all individuals and with GQ values equal to or more than 20 in all individuals. The final dataset consisted of 1,916,277 sites (36 individuals).
Dataset 9: Phylogenetic analyses using three Japanese wolves (Jw225, Jw271, and Jw284), selected dogs (three samples each from dingo/NGSD, African, and sled dogs), modern wolves, and outgroup samples with BQSR (Fig. 2A, Supplementary Figs. 22 and 24)
After exporting bam files, marking duplicated reads, haplotype calling, combining gvcf files, and genotyping, all SNP sites were extracted. The vcf file including SNP sites was filtered by Filtervcf in GATK v4.2 with parameters; “QD<2.0” –filter-name “QD2” -filter “QUAL<30.0” –filter-name “QUAL30” -filter “SOR>4.0” –filter-name “SOR4” -filter “FS>60.0” –filter-name “FS60” -filter “MQ<40.0” –filter-name “MQ40” -filter “MQRankSum<-12.5” –filter-name “MQRankSum-12.5” –filter “ReadPosRankSum<-8.0” –filter-name “ReadPosRankSum-8”. Using the filtered vcf file as “known-sites”, base quality of the bam files of three Japanese wolves (Jw225, Jw271, and Jw284), selected dogs (three samples each from dingo/NGSD, African, and sled dogs), modern wolves, and outgroup samples (Supplementary Data 2) were recalibrated using the BaseRecalibrator and the ApplyBQSR algorithm in GATK v4.2 (BQSR). The recalibrated bam files were genotype called again, and output in gvcf format using the HaplotypeCaller algorithm in GATK v4.2. All gvcf files were combined into a single gvcf format file by the CombineGVCFs algorithm, and a combined file was genotyped by the GenotypeGVCFs algorithm in GATK v4.2. We removed all sites with missing data. Then, we removed singleton and doubleton sites to eliminate PCR and sequencing errors that may have occurred in one individual by minor allele frequency filtering (MAF < 0.035). We extracted bi-allelic sites with coverage equal to or more than three in all individuals and with GQ values equal to or more than 20 in all individuals. The final dataset consisted of 179,397 sites (33 individuals). For a Maximum parsimony tree (Supplementary Fig. 22), we extracted bi-allelic sites with coverage equal to or more than five in all individuals and with GQ values equal to or more than 20 in all individuals without minor allele frequency filtering (the final dataset: 4,864,602 SNPs).
Dataset 10: Phylogenetic analyses using the Japanese wolf (including a Honshu wolf) and modern samples (Supplementary Fig. 13B)
We selected six modern dogs, ancient dogs, four Japanese wolves (Jw255, Jw271, Jw284, and a Honshu wolf), and modern wolves from the genotyped vcf file (Supplementary Data 2). We removed all sites with missing data. Then, we removed all indels, singleton, and doubleton sites to eliminate PCR and sequencing errors that may have occurred in one individual by minor allele frequency filtering (MAF < 0.04). We extracted bi-allelic sites with coverage equal to or more than three in all individuals and with GQ values equal to or more than eight in all individuals. The SNP dataset was pruned and the final dataset consisted of 60,512 sites (36 individuals).
Dataset 11: Phylogenetic analyses using the Japanese wolf (including Jw5k) and modern samples (Supplementary Fig. 13C)
We selected six modern dogs, ancient dogs, four Japanese wolves (Jw255, Jw271, Jw284, and a Jw5k), and modern wolves from the genotyped vcf file (Supplementary Data 2). We removed all sites with missing data. Then, we removed all indels, singleton, and doubleton sites to eliminate PCR and sequencing errors that may have occurred in one individual by minor allele frequency filtering (MAF < 0.04). We extracted bi-allelic sites with coverage equal to or more than three in all individuals and with GQ values equal to or more than eight in all individuals. We extracted transversion sites to eliminate the effect of DNA damage in the ancient samples. The final dataset consisted of 1019 sites (36 individuals).
Dataset 12: Phylogenetic analyses, f3, and f4 statistics using the ancient canids and modern samples (Supplementary Figs. 14A, 15, 17, and 18)
We selected six modern dogs, ancient dogs, eight ancient canids, three Japanese wolves (Jw255, Jw271, and Jw284), and modern wolves from the genotyped vcf file (Supplementary Data 2). We removed all sites with missing data. Then, we removed all indels, singleton, and doubleton sites to eliminate PCR and sequencing errors that may have occurred in one individual by minor allele frequency filtering (MAF < 0.03). We extracted bi-allelic sites with coverage equal to or more than three in all individuals and with GQ values equal to or more than eight in all individuals. We extracted transversion sites to eliminate the effect of DNA damage in the ancient samples. The final dataset consisted of 164,265 sites (43 individuals).
Dataset 13: f4 statistics using the Japanese wolf (including a Honshu wolf) and the ancient canids samples (Supplementary Fig. 14B)
We selected six modern dogs, ancient dogs, eight ancient canids, four Japanese wolves (Jw255, Jw271, Jw284, and a Honshu wolf), and modern wolves from the genotyped vcf file (Supplementary Data 2). We removed all sites with missing data. Then, we removed all indels, singleton, and doubleton sites to eliminate PCR and sequencing errors that may have occurred in one individual by minor allele frequency filtering (MAF < 0.03). We extracted bi-allelic sites with coverage equal to or more than three in all individuals and with GQ values equal to or more than eight in all individuals. We extracted transversion sites to eliminate the effect of DNA damage in the ancient samples. The final dataset consisted of 6190 sites (44 individuals).
Dataset 14: f3 and f4 statistics using the Japanese wolf and the ancient canids (including PJ35k) samples (Supplementary Figs. 14, 16–18)
We selected six modern dogs, ancient dogs, nine ancient canids (including PJ35k), three Japanese wolves (Jw255, Jw271, and Jw284), and modern wolves from the genotyped vcf file (Supplementary Data 2). We removed sites with missingness higher than 20% and minor allele frequency (MAF) < 0.03. We extracted bi-allelic sites with coverage equal to or more than three in all individuals. We extracted transversion sites to eliminate the effect of DNA damage in the ancient samples. The final dataset consisted of 593,0719 sites (44 individuals).
Dataset 15: a species-tree type phylogenetic tree based on coalescent (Supplementary Fig. 23)
We selected individuals listed in Supplementary Data 2 from dataset 1.
Phylogenetic analysis
The SNP dataset 7 was pruned using PLINK ver. 1.941 with an option “–indep-pairwise 50 10 0.1”. The pruned SNP vcf file was converted to PHYLIP format. 10 kb sequences from the 5’ end of the PHYLIP format file were extracted and a model for the Maximum Likelihood method was selected using MEGA ver. X43. A phylogenetic tree was constructed using the Maximum Likelihood (ML) method using PhyML ver. 3.244 with a model selection option “-m GTR” and with 100 bootstrap replications (Supplementary Figs. 19: 327,402 SNPs). The same pruned-vcf file was converted to NEXUS format. A phylogenetic tree was constructed by the svdq algorithm45 in PAUP* ver. 4a (https://paup.phylosolutions.com) with 100 bootstrap replications (Supplementary Fig. 20: 327,402 SNPs).
Using the same pruned-vcf file, we used PLINK ver. 1.941 with an option “—distance 1-ibs” to calculate an Identity By State (IBS) distance matrix using 327,402 SNPs. A neighbour joining tree was constructed from the IBS distance matrix using MEGA ver. X43 (Supplementary Fig. 21).
The SNP datasets 8 and 9 were pruned and a model for the Maximum Likelihood method was selected. Phylogenetic trees for each dataset were constructed using the Maximum Likelihood (ML) method described above (Fig. 2A: 17,489 SNPs and Supplementary Figs. 30 and 31: 157,906 SNPs). A phylogenetic tree for dataset 9 was constructed by the Maximum parsimony method using MEGA ver. X43 (Supplementary Fig. 22) with 1000 bootstrap replications.
We performed StarBeast2 (ver. 1.0.0) to obtain a species-tree type phylogenetic tree based on coalescent46. Because it seems that using every 30 kb window in the genome for the StarBeast2 was practically impossible due to the huge amount of required computation, we decided to sample haploblocks from the genome and use a limited number of samples instead of all the samples we have. The list of samples used (dataset 15) and the regions used are shown in Supplementary Data 2 and 4, respectively.
In order to sample haploblocks, we wish to choose genomic regions within high LD. First, we run plink with “—indep-pairwise 50 10 0.1” option against our genome wide SNPs data as if we are going to do LD pruning. As a result, we obtained a list of SNPs that are in linkage equilibrium. By using this information, we were able to know the region within high LD. Then, we randomly chose 19 regions from those haploblocks longer than 20 kb. We chose one region per one odd number chromosomes to ensure that each region is in linkage equilibrium and that we do not use too many regions for computational ease.
For performing StarBeast2, we used relatively simple models to save computational time. For setting up StarBeast2, we used BEAST software (ver. 2.7.4)47. We used “strict clock model” for all the loci and set the clock as “0.001”. We used HKY for the mutation model setting “Kappa=2” and “empirical.” Population model was set as “constant population.” We ran MCMC for a chain length of 1,000,000,000 to achieve at least 200 of ECC for every parameter to be estimated. We chose “Birth Death Model” for the prior of “Tree.t:Species”. For the other prior, we used the default setting. The output was checked by Tracer ver 1.7.248, and we checked that every parameter has at least 200 ECC. The output trees were visualized by Densitree ver. 2.7.449. In order to obtain the most plausible species tree, “Root Canal” option is used for displaying together with a sampled tree (indicated by a blue tree in Supplementary Fig. 23).
Principal component analysis and ADMIXTURE
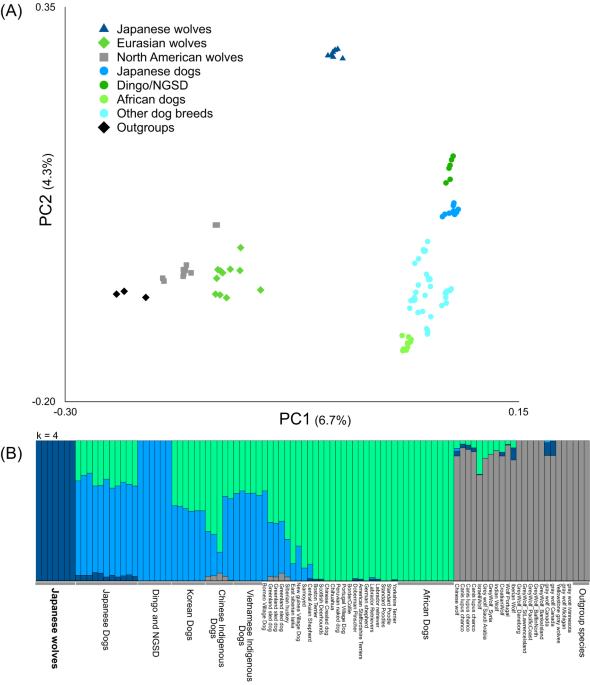
We performed a principal component analysis (PCA) using PLINK ver. 1.941 with an option “–indep-pairwise 50 10 0.1” to explore the affinity among gray wolves, Japanese wolves, and dogs (Fig. 1A). We also performed PCA with type specimens of Japanese wolf (Supplementary Fig. 6), and with ancient canids (Supplementary Figs. 9–12).
ADMIXTURE ver. 1.350 was run on the dataset of modern samples (Fig. 1B and Supplementary Figs. 4 and 5) and modern specimens with type specimens of Japanese wolf (Supplementary Figs. 7, 10–12) assuming 2 to 8 clusters (K = 2–8).
f 3, f4 statistics, and f4-ratio
f 3, f4 statistics, and f4-ratio implemented in ADMIXTOOLS ver. 7.0.128 were used to evaluate the shared genetic drift among gray wolves, Japanese wolves, and dogs using SNP dataset 5 (489,524 SNPs). For Liden b and Leiden c analyses, we prepared two vcf files to maximize the number of SNPs (dataset 6).
Outgroup f3 statistics were calculated to explore shared genetic drift between all dogs and each of the wolves (Fig. 2B), African dogs and each of the wolves (Supplementary Fig. 25A), dingo/NGSD dogs and each of the wolves (Supplementary Fig. 25B), Japanese wolf and each of the dogs (Fig. 2C), dingo/NGSD dogs and each of the other dogs (Supplementary Fig. 35A), and African dogs and each of the other dogs (Supplementary Fig. 35B).
f3 statistics were calculated to test the genomic mixture of African and dingo/NGSD dogs in all dogs (Supplementary Fig. 38).
f4 statistics were calculated to explore shared genetic drift between each of the dogs and Leiden c (Supplementary Fig. 8A: 38,254 SNPs), each of the dogs and Leiden b (Supplementary Fig. 8C: 83,259 SNPs), each of the wolves and Japanese wolves (Fig. 2D), each of the dogs and Japanese wolves (Supplementary Fig. 26), each of the dogs and each of the wolves (Supplementary Fig. 29), dingo/NGSD and Japanese wolves (Supplementary Fig. 33), each of the dogs and Japanese wolves (Supplementary Fig. 34), NGSD1 or Basenji and each of the dogs (Supplementary Fig. 36), and dingoes and each of the other dogs, and African dogs and each of the other dogs (Supplementary Fig. 37).
TreeMix
To examine the admixture events, we used TreeMix ver. 1.1351 to build a tree with admixture edges. We used major groups of gray wolves and dogs as follows; Gray Wolves (North America: n = 2), Gray Wolves (Canada/Arctic: n = 8), Gray Wolves (East Asia: n = 5), Gray Wolves (West Eurasia: n = 7), Japanese Wolves (Japan: n = 7), Dogs (Central Asian: n = 4), Dogs (Europe: n = 11), Dogs (Africa: n = 10), Dogs (sled dogs: n = 4), Dogs (Vietnamese Indigenous: n = 5), Ding/NGSD (Oceania: n = 7), Japanese Dogs (Japan: n = 11), and Dogs (Korea: n = 6). The SNP dataset was pruned based on linkage disequilibrium (LD-pruning) by using plink with an option “–indep-pairwise 50 10 0.12”. As a result of LD-pruning, 150,502 SNPs were used for TreeMix.
Assessing DNA damage patterns
We used mapDamage ver. 2.2.052 to assess DNA damage patterns in the Japanese wolf samples sequenced in this study. Mapped reads from the Japanese wolf samples showed slightly increased proportion (equal to or less than 1%) of C to T and G to A substitutions at the 5’ and 3’ read ends, respectively (Supplementary Fig. 1).
Calculation of maximum contamination rate
We used substitutions in mitochondria DNA specific to Japanese wolf to assess the contamination rate of the other animal DNA in Japanese wolf DNA. Fifteen fixed substitutions unique to Japanese wolf were selected using an alignment of mitochondria DNA sequences including gray wolves, Japanese wolves, and dogs used in previous studies25,53. The lowest coverage at fifteen sites was 48 (highest was 28,324) in the mapping result to the mitochondria genome in CanFam3.1. We calculated the average mapping ratio in fifteen sites. The ratio of the reads mapped to the fifteen sites without substitutions specific to the Japanese wolf were assumed as the maximum contamination rate (Supplementary Data 5), because the mitochondria DNA-like sequences are found in the nuclear genome.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.